Saturday, February 28, 2015
Levenshtein Distance in R
Levenshtein distance is a measure of how many characters should be replaced or moved to get two strings same.
In the example below, a string text is asked from the user in console mode. Then the input string is compared to colour names defined in R. Similar colour names are then reported:
user.string <- readline("Enter a word: ")
wordlist <- colours()
dists <- adist(user.string, wordlist)
mindist <- min(dists)
best.ones <- which(dists == mindist)
for (index in best.ones){
cat("Did you mean: ", wordlist[index],"\n")
}
Here is the results:
Enter a word: turtoise
Did you mean: turquoise
Enter a word: turtle
Did you mean: purple
Enter a word: night blue
Did you mean: lightblue
Enter a word: parliament
Did you mean: darkmagenta
Enter a word: marooon
Did you mean: maroon
Have a nice read
Friday, February 27, 2015
Frequency table of characters in a string in R
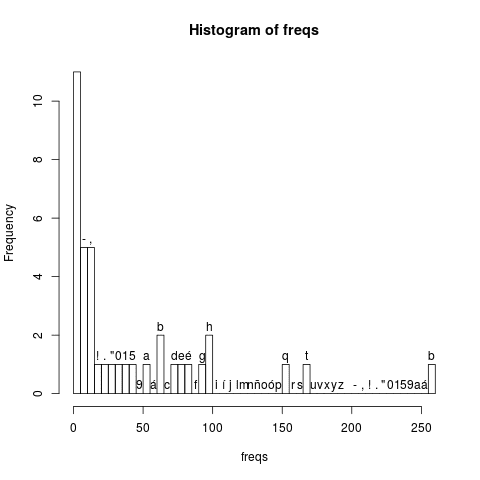
Here is a example on a text that is captured from the Oracle - History of Java site. The code below defines a large string. The string is then parsed into its characters. After calculating the frequencies of each single character (including numbers, commas and dots) a histogram is saved in a file.
# Defining string
s <- "Since 1995, Java has changed our world and our expectations. Today, with technology such a part of our daily lives, we take it for granted that we can be connected and access applications and content anywhere, anytime. Because of Java, we expect digital devices to be smarter, more functional, and way more entertaining. In the early 90s, extending the power of network computing to the activities of everyday life was a radical vision. In 1991, a small group of Sun engineers called the \"Green Team\" believed that the next wave in computing was the union of digital consumer devices and computers. Led by James Gosling, the team worked around the clock and created the programming language that would revolutionize our world – Java. The Green Team demonstrated their new language with an interactive, handheld home-entertainment controller that was originally targeted at the digital cable television industry. Unfortunately, the concept was much too advanced for the them at the time. But it was just right for the Internet, which was just starting to take off. In 1995, the team announced that the Netscape Navigator Internet browser would incorporate Java technology.Today, Java not only permeates the Internet, but also is the invisible force behind many of the applications and devices that power our day-to-day lives. From mobile phones to handheld devices, games and navigation systems to e-business solutions, Java is everywhere!"
# First converting to lower case
# then splitting by characters.
# strsplit return a list, we are unlisting to a vector.
chars <- unlist(strsplit(tolower(s), ""))
# Generating frequency table
freqs <- table(chars)
# Generating plot into a file
png("Graph.png")
hist(freqs,include.lowest=TRUE, breaks=46,freq=TRUE,labels=rownames(freqs))
dev.off()
The generated output is
We translate the text used in our example to Spanish using Google Translate site. The code is shown below:
# Defining string
s <- "Desde 1995, Java ha cambiado nuestro mundo y nuestras expectativas. Hoy en día, con la tecnología de una parte de nuestra vida cotidia na tal, damos por sentado que se puede conectar y acceder a las aplicaciones y contenido en cualquier lugar ya cualquier hora. Debido a Java , esperamos que los dispositivos digitales para ser más inteligente, más funcional, y de manera más entretenida. A principios de los años 90 , que se extiende el poder de la computación en red para las actividades de la vida cotidiana era una visión radical. En 1991, un pequeño gr upo de ingenieros de Sun llamado \"Green Team\" cree que la próxima ola de la informática fue la unión de los dispositivos digitales de cons umo y ordenadores. Dirigido por James Gosling, el equipo trabajó durante todo el día y creó el lenguaje de programación que revolucionaría e l mundo - Java. El Equipo Verde demostró su nuevo idioma con una mano controlador interactivo, el entretenimiento en casa que fue dirigido o riginalmente a la industria de la televisión digital por cable. Por desgracia, el concepto fue demasiado avanzado para el ellos en el moment o. Pero fue justo para Internet, que estaba empezando a despegar. En 1995, el equipo anunció que el navegador de Internet Netscape Navigator incorporaría Java technology.Today, Java no sólo impregna el Internet, pero también es la fuerza invisible detrás de muchas de las aplicaci ones y dispositivos que alimentan nuestra vida del día a día. Desde teléfonos móviles para dispositivos de mano, juegos y sistemas de navega ción para e-business soluciones, Java está en todas partes!"
# First converting to lower case
# then splitting by characters.
# strsplit return a list, we are unlisting to a vector.
chars <- unlist(strsplit(tolower(s), ""))
# Generating frequency table
freqs <- table(chars)
# Generating plot into a file
png("Graph.png")
hist(freqs,include.lowest=TRUE, breaks=46,freq=TRUE,labels=rownames(freqs))
dev.off()
Environments in R
After setting a variable to a value out of any function and class, the default holder of this variable is the global environment.
Suppose we set t to 10 by
t <- 10
and this is the same as writing
assign(x="t", value=12, envir=.GlobalEnv)
and the value of t is now 12:
> t <- 10
> assign(x="t", value=12, envir=.GlobalEnv)
> t
[1] 12
Instead of using the global environment, we can create new environments and attach them to parent environments. Suppose we create a new environment as below:
> my.env <- new.env()
> assign(x="t", value="20", envir=my.env)
> t
[1] 12
Variables in environments are accessable using the assign and the get functions.
> get(x="t", envir=my.env)
[1] "20"
> get(x="t", envir=.GlobalEnv)
[1] 12
> my.env <- as.environment(my.list)
> get("a", envir=my.env)
[1] 3
> get("b", envir=my.env)
[1] 7
Wednesday, February 18, 2015
Fast and robust estimation of regression coefficients with R
Since an outlier may change the partial coefficients of regression, examining the residuals of a non-robust estimator results wrong conclusions. An outlier may change one or more regression coefficients and hide itself with a relatively small residual. This effect is called masking. This change in coefficients can get a clean observation distant from the regression object with higher residual. This effect is called swamping. A successful robust estimator should minimize these two effects to estimate regression coefficients in more precision.
The medmad function in R package galts can be used for robust estimation of regression coefficients. This package is hosted in the CRAN servers and can be installed in R terminal by typing
install.packages("galts")
Once the package is installed, its content can be used by typing
require("galts")
and the functions and help files can be ready to use after typing an enter key. Here is a complete example of generating a regression data, contaminating some observations and estimating the robust regression coefficients:
The output is
(Intercept) x1 x2
4.979828 4.993914 4.985901
in which the parameters are near to 5 as the data is generated before. The details of this algorithm can be found in the paper
Satman, Mehmet Hakan. "A New Algorithm for Detecting Outliers in Linear Regression." International Journal of Statistics and Probability 2.3 (2013): p101.
which is avaliable at site
http://www.ccsenet.org/journal/index.php/ijsp/article/view/28207
and
http://www.ccsenet.org/journal/index.php/ijsp/article/download/28207/17282
Have a nice detect!
Tuesday, April 22, 2014
Word Cloud Generation Using Google Webmaster Tools Data and R
Google Index -> Content keyword
you get the keywords with their frequencies. This data set can be saved in csv format using the button "Download this table".
The csv data for our blog was like this:
One can load this file and generate a word cloud graphics using R, and our code is shown below:
The generated output is:
Here is the stdioe's search query keywords cloud:
Monday, April 21, 2014
Matrix Inversion with RCaller 2.2
RCaller caller = new RCaller(); Globals.detect_current_rscript(); caller.setRscriptExecutable(Globals.Rscript_current); RCode code = new RCode();
double[][] matrix = new double[][]{{6, 4}, {9, 8}}; code.addDoubleMatrix("x", matrix); code.addRCode("s<-solve font="" x="">); caller.setRCode(code); caller.runAndReturnResult("s"); double[][] inverse = caller.getParser().getAsDoubleMatrix("s",
matrix.length, matrix[0].length);
for (int i = 0; i < inverse.length; i++) { for (int j = 0; j < inverse[0].length; j++) {
System.out.print( inverse[i][j] + " ");
}
System.out.println(); }
Tuesday, August 20, 2013
A gWidgets Example - Using windows, groups, labels, text and password boxes, buttons and events in R
A gWidget Example - Using windows, groups, labels, text and password boxes, buttons and events in R
In this entry, a short example for using gWidgets is given. gWidgets is a package for creating GUI’s in R.
Our example shows a GUI window with width = 400 and height = 400. Window is created by gwindow function. Components are located by rows. Rows are handled by ggroup function. ggroup must take a container as a parameter. In this logic, lbl_username and txt_username are childs of row1 which is child of gwindow.
Any text field can act as a password field by using visible¡- function.
So, the object txt_password is now hiding characters by * characters. Finally, the method addHandlerClicked links an object to a function for click event. In our example, btn_login is linked to do_login function. When btn_login clicked, a message is written. The source code of the complete example is given below.
1# Loading required packages
2require("gWidgets")
3require("gWidgetstcltk")
4
5# main window
6main <- gwindow(title="Login␣Window",
7 width=400,
8 height=400)
9
10# a row and components
11row1 <- ggroup(container=main)
12lbl_username <- glabel(container=row1, text="Username:␣")
13txt_username <- gedit(container=row1)
14
15
16# a row and components
17row2 <- ggroup(container=main)
18lbl_password <- glabel(container=row2, text="Password:␣")
19txt_password <- gedit(container=row2)
20
21# any text in txt_password will be show with * character
22visible(txt_password) <- FALSE
23
24# a row for button
25row3 <- ggroup(container=main)
26btn_login <- gbutton(container=row3,
27 text="Login")
28btn_register <- gbutton(container=row3,
29 text="Register")
30
31
32# Event handler for login button
33do_login <- function(obj){
34 cat("Login␣with␣",svalue(txt_username),"\n")
35}
36
37# Event handler for register button
38do_register <- function(obj){
39 cat("Register␣with␣", svalue(txt_username),"\n")
40}
41
42
43# Registering Events
44addHandlerClicked ( btn_login, do_login)
45addHandlerClicked ( btn_register, do_register)